Genetic selection of Calophyllum brasiliense for seed orchards
DOI:
https://doi.org/10.20873/jbb.uft.cemaf.v4n4.schuhliKeywords:
RAPD, propagative material, MAS, guanandiAbstract
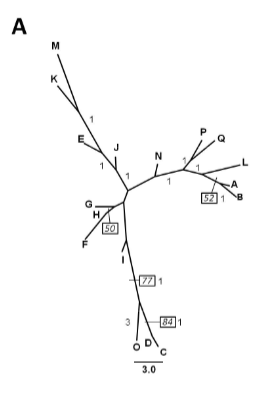
Calophyllum brasiliense populations are under severe depletion and criteria to improve production and quality of propagative material are therefore necessary. Genetic measures have the potential to reduce consanguinity and maximize allelic representation within target populations. Here, we explored genetic values for this species in a small relic of natural forest in Rio de Janeiro State (Brazil). The objective was to evaluate the potential of some genetic measures for seed orchard establishment. As genomic information of native trees is still scarce, we opted to use a dominant marker: RAPD. DNA from 17 phenotypically superior trees was obtained through the CTAB method and submitted for amplification by PCR. Electrophoresis and electronic documentation was then conducted. We calculated the percentage of polymorphic bands (PPB), gene diversity (Ht), Shannon’s information index (i), genetic distance (UPGMA) and parsimony analysis. Six primers were evaluated generating 34 loci. We found high genetic diversity PPB=70.6% with Ht=0.28 and i=0.41. Genetic relationships were reported in dendrograms (maximum parsimony and distance). Simulated sampling within and among clusters suggests that inter cluster sampling is more effective to capture the genetic diversity.
References
Bremer K. Branch support and tree stability. Cladistics. 1994; 10: 295-304.
Caldas L. Pomares de sementes de espécies nativas – As funções das redes de sementes. In: Higa AR. and Silva LD. Pomar de sementes de espécies florestais nativas. 1st ed. FUPEF; 2006. 227-242.
Cheung WY, Hubert N, Landry BS. A simple and rapid DNA microextration method for plant, animal, and insect suitable for RAPD and other PCR analyses. Genome Res. 1993; 3: 69-70.
Cruz CD. Programa Genes: Biometria. Viçosa (MG): Editora UFV; 2006.
Kruskal JB. Multidimensional scaling by optimizing goodness of fit to a no metric hypothesis. Psychometrika. , 1964; 29(1): 1-27.
Lazar I. [homepage on the internet].Gel Analyzer. Debrecen, Hungary: University of Debrecen; updated in 2010; accessed in 2013 Jan 11. Available from: http://www.gelanalyzer.com/index.html
Manly BFJ. Randomization, bootstrap and Monte Carlo methods in Biology. London: Chapman & Hall, 1997.
Marques MCM, Joly CA. Estrutura e dinâmica de uma população de Calophyllum brasiliense Camb. em floresta higrófila do sudeste do Brasil. Rev Bras Bot. 2000; 23(1):107-12.
Muchugi A, Kadu C, Kindt R, Kipruto H, Lemurt, S, Olale K, Nyadoi P, Dawson I, Jamnadass, R. Molecular markers for tropical trees: A Practical guide to principles and procedures. ICRAF Technical Manual no. 9. World Agroforestry Centre Nairobi; 2009.
Nei M, Li W-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA. 1979; 76(10): 5269-73.
Nei M. Genetic distance between populations. Am. Nat. 1972; 106:283-92.
Newbury HJ, Ford-Lloyd BV. The use of RAPD for assessing variation in plants. Plant Growth Regul. 1993; 12:43-51, 1993.
Oliveira-Filho AT, Ratter JA. A study of the origin of central brazilian forests by the analysis of plant species distribution patterns. Edinb J Bot. 1995; 52: 141-94.
Reitz R, Klein RM, Reis A. Projeto madeira de Santa Catarina. Sellowia. 1978; 28(30): 218-24.
Rohlf FJ. NTSYS-pc v. 1.8. Numerical taxonomy and multivariate analysis system. Setauket, NY: Applied Biostatistics Inc. 1993.
Sebbenn, AM. Número de árvores matrizes e conceitos genéticos na coleta de sementes para reflorestamentos com espécies nativas. Rev Inst Flor. 2002; 14: 115-132.
Sebbenn, AM. Sistemas de reprodução em espécies arbóreas tropicais e suas implicações para a seleção de árvores matrizes para reflorestamentos ambientais. In: Higa AR. and Silva LD. Pomar de sementes de espécies florestais nativas. 1st ed. FUPEF; 2006. 93-138.
Swofford, D. L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4b10. Sunderland, Massachusetts: Sinauer Associates. 2003.
Tivang JG, Nienhuis J, Smith OS. Estimation of sampling variance of molecular marker data using the bootstrap procedure. Theor Appl Genet. 1994; 89(25):264-9.
Yeh F, Yang RC, Mao J, Ye Z, Boyle TJB. The user-friendly shareware for Population Genetic Analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton, 1999.
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 - Journal of Biotechnology and Biodiversity

This work is licensed under a Creative Commons Attribution 4.0 International License.
Authors who publish with this journal agree to the following terms:
Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License (CC BY 4.0 at http://creativecommons.org/licenses/by/4.0/) that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.
Authors are permitted and encouraged to post their work online (e.g. in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (Available at The Effect of Open Access, at http://opcit.eprints.org/oacitation-biblio.html).


