Genetic Variability of Guzerat and Senepol Bovine Breeds by Microsatellite Markers
DOI:
https://doi.org/10.20873/jbb.uft.cemaf.v4n3.silvafilhoPalabras clave:
Cattle, STRs, diversity, livestock conservationResumen
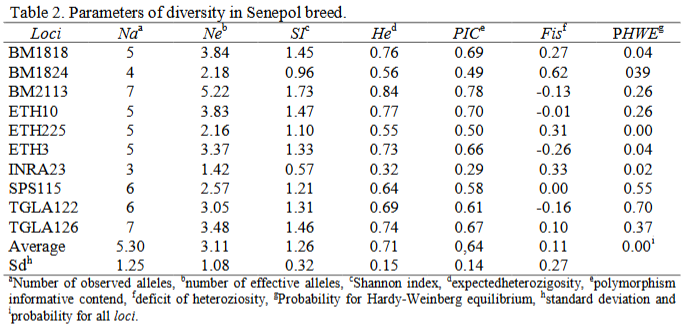
Bovine production plays economic importance in Brazil and Guzerat and Senepol breeds are producer of meat. It was aimed to analyze the genetic variability of Guzerat and Senepol breeds by microsatellite markers. The breeds were collected and genotyped for ten microsatellite loci by automatic sequencer and statistically analysed. A total of 53 alleles were observed being the average number was 5.3 in both breeds. The effective numbers of alleles were 3.36 for Guzerat and 3.11 for Senepol cattle. The Shannon indexes were 1.36 for Guzerat and 1.26 for Senepol cattle. The expected heterozigosity were 0.71 and PIC values were 0.64 in both breeds. The FIS were 0.01 and 0.11 for Guzerat and Senepol breeds, respectively and Hardy-Weinberg equilibrium were P>0.05 for Guzerat and P<0.05 for Senepol cattle. The combined discrimination powers were 0.99 in both breeds and combined exclusion powers (PE1) were 0.99 in both breeds and combined exclusion powers (PE2) were 0.96 and 0.95 for Guzerat and Senepol breeds espectively. There is genetic variability in both breed, but there are evidences of inbreeding enabling genetic drift and should be necessary to use major number of microsatellite loci to analyze with high efficiency the exclusion power (PE2).
Citas
ARIF, I.A.;KHAN, H.A.;SHOBRAK, M.;HOMAIDAN, A.A.;SADOON, M.A.;FARHAN, A.H. Measuring the genetic diversity of Arabian Oryx using microsatellite markers: implication for captive breeding. Genes & Genetic Systems, v.85, p.141-145, 2010.
BOTSTEIN, D.; WHITE, R.L.; SKOLNICK, M.; DAVIS, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics, v.32, p.314- 331, 1980.
CARNEIRO, T.X.; GONÇALVES, E.C.; SCHNEIDER, M.P.C.; SILVA, A. Diversidade genética e eficiência de DNA microssatélites para o controle genealógico da raça Nelore. Arquivo Brasileiro de Medicina Veterinária e Zootecnia, v.59, p.1257-1262, 2007.
CERVINI, M.; SILVA, H.F.; MORTARI, N.; MATHEUCCI, J.R.E. Geneticvariabilityof 10 microsatellitemarkers in thecharacterizationofBrazilianNellorecattle (Bosindicus). Genetics and Molecular Biology, v.29, p.486-490, 2006.
CHARLESWORTH, B.; SNIEGOWSKI, P.; STEPHAN, W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature, v.371, p.215- 220, 1994.
DALVIT, C.; DE MARCHI, M.; DAL ZOTTO, R.; ZANETTI, E.;MEUWISSEN, T.; CASSANDRO, M. Genetic characterization of the Burlina cattle breed using microsatellites markers. Journal of Animal Breeding and Genetics, v.125, p.137–144, 2008.
EDWARDS, A.;CIVITELLO, A.;HAMMOND, H.A.;CASKEY, C.T. DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. American Journal of Human Genetics, v.49, p.746-756, 1991.
EDWARDS, A.; HAMMOND, H.A.; JIM, L. et al.Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics, v.12, p.241-253, 1992.
EGITO, A.A.; PAIVA, S.R.; ALBUQUERQUE, M.D.S.M.; MARIANTE, A.S.; ALMEIDA, L.D.; CASTRO, S.R.; GRATTAPAGLIA, D.Microsatellite based genetic diversity and relationships among ten Creole and commercial cattle breeds raised in Brazil. BMC Genetics, v.8, p.1-14, 2007.
FATIMA, S.;BHONG, C.D.;RANK, D.N.;JOSHI, C.G. Genetic variability and bottleneck studies in Zalawadi, Gohilwadi and Surti goat breeds of Gujarat (India) using microsatellites. Small Ruminant Research, v.77, p.58-64, 2008.
FISHER, R. Standard calculations for evaluating a blood group system. Heredity, v. 5, p. 95-102, 1951. IBGE, Instituto Brasileiro de Geografia e Estatística. Produção da Pecuária Municipal, Rio de Janeiro, RJ, Brasil, v.39, 2011.63p.
JAMIESON, A.; TAYLOR, S.C. Comparisons of three probability formula for parentage exclusion. Animal Genetics, v.28, p.397-400, 1997.
KANTANEN, J., OLSAKER, I., HOLM, L.E.; LIEN, S.; VILKKI, J.; BRUSGAARD, K.; EYTHORSDOTTIR, E.; DANELL, B.; ADALSTEINSSON, S. Genetic diversity and population structure of 20 north European cattle breeds. The Journal of Heredity, v.91, p.446-457, 2000.
KARTHICKEYAN, S.M.K.; SIVASELVAM, S.N.; SELVAM, R.; THANGARAJU, P. Microsatellite analysis of Kangayam cattle (Bosindicus) of Tamilnadu.Indian Journal of Science and Technology, v.2, p.38-40, 2009.
KLOOSTERMAN, A.D.; BUDOWLE, B.; DASELAAR, P. PCR-amplification and detection of the human D1S80 VNTR locus, Amplification conditions, population genetics and application in forensic analysis. International Journal of Legal Medicine, v.7, p.257-264, 1993.
NEY, M.; ROYCHOUDHURY, A.K. Sampling variances of heterozygosity and genetic distance. Genetics, v.76, p.379-390, 1974.
PAETKAU, D.; CALVERT, W.; STIRLING, I.; STROBECK, C. Microsatellite analysis of population structure in Canadian polar bears. Molecular Ecology, v.4, p.347-354, 1995.
RAYMOND, M.; ROUSSET, F. GENEPOP (Version 1.2): Population Genetics Software for Exact Tests and Ecumenicism. Journal of Heredity, v.86, p.248-249, 1995.
REHMAN, M.S.; KHAN, M.S. Genetic diversity of hariana and hissar cattle from pakistan using microsatellite analysis. Pakistan Veterinary Journal, 29, 67-71, 2009.
ROY, M.S., GEFFEN, E., SMITH, D.; OSTRANDER, E.A.; WAYNE, R.K.Patterns of differentiation and hybridization in North American wolflikecanids, revealed by analysis of microsatellite loci.Molecular Biology and Evolution, v.11, p.553-570, 1994.
RUBINSZTEIN, D.C. Trinucleotide expansion mutations cause diseases which do not conform to classical Mendelian expectations. In: Goldstein, D.B.;Schlötterer, C. (eds) Microsatellites: evolution and applications. Oxford University Press, 1999, pp 80–97.
SAMBROOK, J.; FRITSCH, E.F.; MANIATIS, T. Molecular Cloning. In A Laboratory Manual. Cold Spring Harbor Laboratory Press. New York, v. 1, 2 and 3. 1989.
SERRANO, G.M.; EGITO, A.A.; MCMANUS, C.; MARIANTE, A.S. Genetic diversity and population structure of Brazilian native bovine breeds. PesquisaAgropecuáriaBrasileira, v.39, p.543-549, 2004.
SHEKAR, M.C.; KUMARA, J.U.; KARTHICKEYAN, S.M.K.; MUTHEZHILAN, R. Assessment of with-in breed diversity in Hallikar cattle (Bosindicus) through microsatellite markers. IndianJournalof Science and Technology, v.4, p.895- 898, 2011.
SILVA FILHO, E.; SCHNEIDER, M.P.C.; SILVA, A. Variabilidade Genética de Cavalos baseada em DNA Microssatélites. RevistaTrópica: CiênciasAgrárias eBiológicas, v.1, p.76-87, 2007.
STEVANOVIC, J.; STANIMIROVIC, Z.; DIMITRIJEVIC, V.; MALETIC, M. Evaluation of 11 microsatellite loci for their use in paternity testing in Yugoslav Pied cattle (YU Simmental cattle).Czech Journal of Animal Science, v.55, p.221–226, 2010.
WEIR, B.S.; COCKERHAM, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution, v.38, p.1358-1370, 1984.
WEISSENBACH, J., GYAPAY, G., DIB, C.; VIGNAL, A.; MORISSETTE, J.; MILLASSEAU, P.; VAYSSEIX, G.; LATHROP, M. A second-generation linkage map of the human genome. Nature, v.359, p.794–801, 1992.
YEH, F.; YANG, C.; BOYLE, T. POPGENE version 1.32 Microsoft window-based freeware for Population Genetic Analysis. University of Alberta. Edmonton, AB. Canada. 1999.
Publicado
Cómo citar
Número
Sección
Licencia
Copyright (c) 2024 - Journal of Biotechnology and Biodiversity

Esta obra está bajo una Licencia Creative Commons Atribución 4.0 Internacional.
Los autores que publican en esta revista aceptan los siguientes términos:
Los autores mantienen los derechos autorales y conceden a la revista el derecho de primera publicación, con el trabajo simultáneamente licenciado bajo la LicenciaCreative Commons Attribution (CC BY 4.0 en el link http://creativecommons.org/licenses/by/4.0/) que permite compartir el trabajo con reconocimiento de la autoría y publicación inicial en esta revista.
Los autores tienen autorización para asumir contratos adicionales separadamente, para distribución no exclusiva de la versión del trabajo publicado en esta revista (ej.: publicar en repositorio institucional o como capítulo de libro), con reconocimiento de autoría y publicación inicial en esta revista.
A los autores se les permite, y son estimulados, a publicar y distribuir su trabajo online (ej.: en repositorios institucionales o en su página personal) en cualquier punto antes o durante el proceso editorial, ya que esto puede generar alteraciones productivas, bien como aumentar el impacto y la citación del trabajo publicado (disponible en El Efecto del Acceso Libre en el link http://opcit.eprints.org/oacitation-biblio.html).


